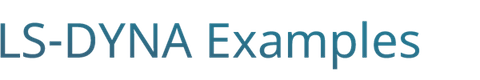Advanced : Centrifugal Blood Pump Flow
*KEYWORD *TITLE *DATABASE_BINARY_D3PLOT *DEFINE_CURVE_TITLE *ICFD_BOUNDARY_NONSLIP
*ICFD_BOUNDARY_PERIODIC *ICFD_BOUNDARY_PRESCRIBED_PRE *ICFD_BOUNDARY_PRESCRIBED_VEL
*ICFD_CONTROL_IMPOSED_MOVE *ICFD_CONTROL_MESH *ICFD_CONTROL_OUTPUT
*ICFD_CONTROL_TAVERAGE *ICFD_CONTROL_TIME
*ICFD_CONTROL_TURBULENCE
*ICFD_DATABASE_DRAG
*ICFD_DATABASE_FLUX *ICFD_MAT *ICFD_PART *ICFD_PART_VOL *ICFD_SECTION *INCLUDE
*MESH_BL
*MESH_SIZE_SHAPE *MESH_SURFACE_ELEMENT *MESH_SURFACE_NODE *MESH_VOLUME *PART *SECTION_SHELL *END
$-----------------------------------------------------------------------------
$
$ Example provided by Peggy and Iñaki (LST-Ansys)
$
$
$X------------------------------------------------------------------------------
$X
$X 1. Run file as is.
$X Requires LS-DYNA MPP R12.0.0 (or higher) with double precision
$X
$X------------------------------------------------------------------------------
$# UNITS: (S.I)
$X------------------------------------------------------------------------------
$X
*KEYWORD
*INCLUDE
mesh.k
*PARAMETER
$--- Fluid
$
R v_inlet 0.3638
Rrho_fluid 1035.0
R mu_fluid 0.0035
R dt_fluid 1.0e-5
R turb_I 0.07
$
$---- Rotor
$
R omega 366.52
$
$ ----------------- Control General --------------------------------------------
*ICFD_CONTROL_OUTPUT
$# msgl
4
*ICFD_CONTROL_TURBULENCE
3
*ICFD_CONTROL_MESH
1.2
*ICFD_CONTROL_TIME
$# ttm dt cfl lcidsf dtmin dtmax dtinit tdeath
0.34288 0 1.0 0 0.0 0.0 0.0 0.0
$ ----------------- Control Sliding --------------------------------------------
*ICFD_BOUNDARY_PERIODIC
4,3,5,
*ICFD_CONTROL_IMPOSED_MOVE
15
,,,100
*ICFD_CONTROL_IMPOSED_MOVE
5
,,,100
*ICFD_CONTROL_IMPOSED_MOVE
6
,,,100
$ ----------------- Parts and Boundary conditions-------------------------------
*ICFD_BOUNDARY_NONSLIP
$# pid
3
6
*ICFD_BOUNDARY_PRESCRIBED_VEL
$# pid dof vad lcid sf vid death birth
1 4 1 20 1.0 01.00000E28 0.0
*ICFD_BOUNDARY_PRESCRIBED_PRE
$# pid lcid sf death birth
2 2 1.01.00000E28 0.0
*ICFD_MAT
$# mid flg ro vis st
1 1&rho_fluid &mu_fluid 0.0
*ICFD_PART_TITLE
inlet
$# pid secid mid
1 1 1
*ICFD_PART_TITLE
outle
$# pid secid mid
2 1 1
*ICFD_PART_TITLE
rotor
$# pid secid mid
6 1 1
*ICFD_PART_TITLE
housing
$# pid secid mid
3 1 1
*ICFD_PART_TITLE
sliding_out
$# pid secid mid
4 1 1
*ICFD_PART_TITLE
sliding_in
$# pid secid mid
5 1 1
*ICFD_PART_VOL_title
fluid_volume_out
$# pid secid mid
15 1 1
5,6
*ICFD_PART_VOL_TITLE
fluid_volume_in
$# pid secid mid
16 1 1
1,2,3,4
*ICFD_SECTION
$# sid
1
$-----------------------------Output--------------------------------------------
*ICFD_CONTROL_TAVERAGE
$# dt
0.017143
*ICFD_DATABASE_FLUX
1
*ICFD_DATABASE_FLUX
2
*ICFD_DATABASE_DRAG
6
*DATABASE_BINARY_D3PLOT
$# dt lcdt beam npltc psetid
0.017143
$# ioopt
0
$-----------------------------MESH----------------------------------------------
*MESH_VOLUME
$# volid
15
$# pid1 pid2 pid3 pid4 pid5 pid6 pid7 pid8
5,6
*MESH_VOLUME
$# volid
16
$# pid1 pid2 pid3 pid4 pid5 pid6 pid7 pid8
1,2,3,4
*MESH_BL
$# pid nelth blth blfe blst
3 4
*MESH_BL
$# pid nelth
6 4
*MESH_SIZE_SHAPE
$# sname
box
$# msize pminx pminy pminz pmaxx pmaxy pmaxz
0.00035 0.0 -0.03 0.004 0.024 -0.026 0.009
$ ----------------- Define Curves ----------------------------------------------
*DEFINE_CURVE
$# lcid sidr sfa sfo offa offo dattyp lcint
1 0 1.0 1.0 0.0 0.0 0 0
$# a1 o1
0.0 1.0
10000.0 1.0
*DEFINE_CURVE
$# lcid sidr sfa sfo offa offo dattyp lcint
2 0 1.0 1.0 0.0 0.0 0 0
$# a1 o1
0.0 0.0
10000.0 0.0
*DEFINE_CURVE
$# lcid sidr sfa sfo offa offo dattyp lcint
3 0 1.0 &turb_I 0.0 0.0 0 0
$# a1 o1
0.0 1.0
10000.0 1.0
*DEFINE_FUNCTION
20,x-velo
f(y,z)=-4.1977e14*(y**2+(z-0.294)**2)**3+5.5e12*sqrt(y**2+(z-0.294)**2)**5
-2.65e10*(y**2+(z-0.294)**2)**2+5.64e7*sqrt(y**2+(z-0.294)**2)**3
-5.86e4*(y**2+(z-0.294)**2)+2.365*sqrt(y**2+(z-0.294)**2)+1.157
*DEFINE_CURVE_TITLE
Rotation velocity
$# lcid sidr sfa sfo offa offo dattyp
100 &omega
$# a1 o1
0.0 0.0
0.0857125 1.0
10000.0 1.0
*END
A key step in the development of medical devices is the testing phase. Regulatory agencies like the Food and Drug Administration require extensive laboratory testing and a long, tedious and expensive clinical trial process before a device is approved for clinical use. In an effort to optimize this process, the industry and the regulatory agencies are looking at numerical methods as an additional tool that could potentially reduce the time of product development, animal testing and cost.
The first critical step is to increase the confidence, reliability and robustness of numerical techniques. The FDA is actively working on this subject by designing laboratory experiments that could be used to evaluate the solution accuracy provided by computational methods launching the Critical Path Initiative (CPI) [1] program. The aim of the program is to standardize the use of computational simulation on the design of the blood-contacting medical devices and analysis of the ratio of hemolysis in them. The goal of this project is to establish the guidelines for applying Computational Fluid Dynamics (CFD) techniques on the evaluation and the optimization of the medical devices.
Among the benchmark proposed ([2]) is the analysis of the flow in a simplified centrifugal blood pump which has been the subject of investigations using the ICFD solver in Huang [3].
The flow enters the chamber through a curved tube with diameter 12 mm. The diameter of the inner chamber of the housing is 60 mm and the thickness is 9 mm. The rotor inside the chamber has a diameter of 52 mm and 4 mm thick, along with four 3 mm thick straight blades. The chamber is connected with a throat at its outlet, followed by a diffuser to the outlet tube with diameter 12 mm. The flowrate is Q = 6 L/min and the rotational speed 3500 RPM.
[1] FDA Critical Path Initiative (CPI), 2016. https://www.fda.gov/scienceresearch/specialtopics/criticalpathinitiative/default.htm
[2] FDAs \Critical Path" Computational Fluid Dynamics (CFD)/Blood Damage Project: Computational Round Robin problems, 2004. https://nciphub.org/wiki/FDA CFD
[3] Huang, CJ (2019). “On the performance and accuracy of PFEM-2 in the solution of biomedical benchmarks”. Computational Particle Mechanics volume 7, pages121–138(2020).